Identificación de péptido antimicrobiano a partir de manzanilla romana
Cargando...
Autores
Tipo de contenido
Document language:
Español
Fecha
Título de la revista
ISSN de la revista
Título del volumen
Imagenes y Videos
Slide 1 of 27 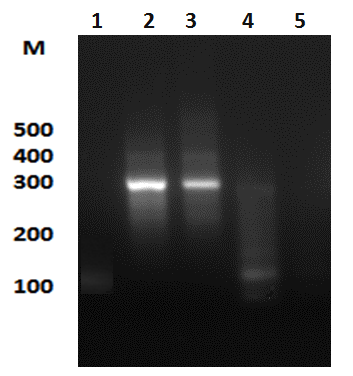
image6.png
Slide 2 of 27 
image7.png
Slide 3 of 27 
image24.png
Slide 4 of 27 
image17.png
Slide 5 of 27 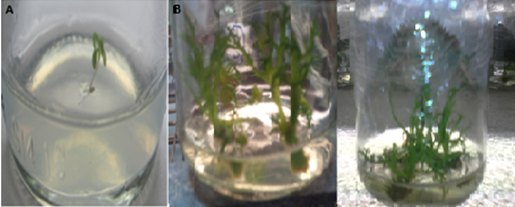
image5.png
Slide 6 of 27 
image23.png
Slide 7 of 27 
image9.png
Slide 8 of 27 
image4.png
Slide 9 of 27 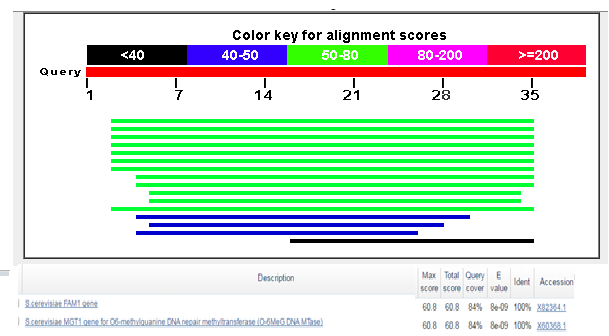
image20.png
Slide 10 of 27 
image25.png
Slide 11 of 27 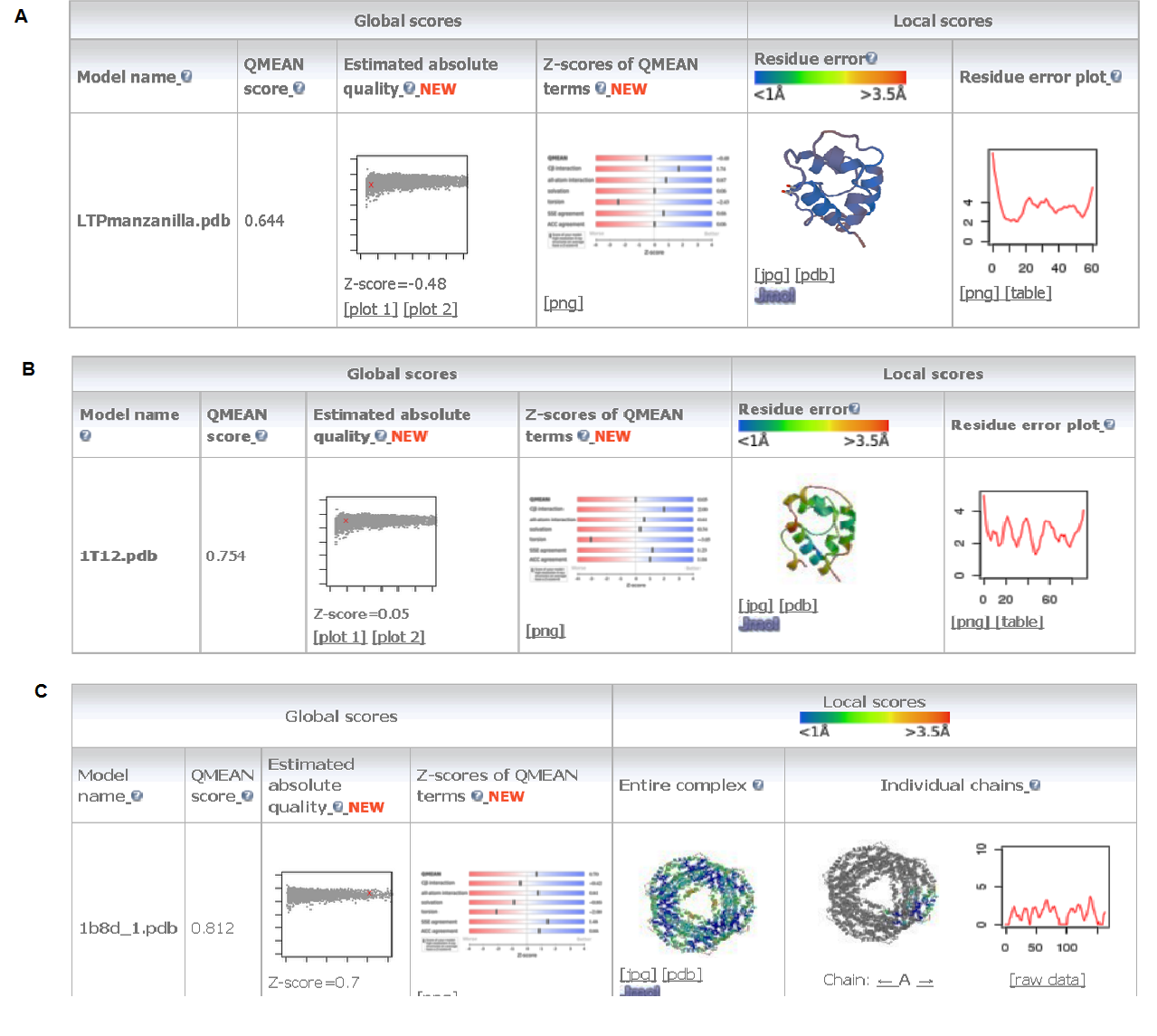
image13.png
Slide 12 of 27 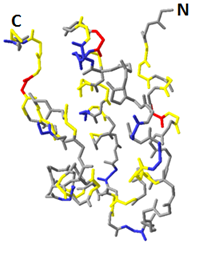
image11.png
Slide 13 of 27 
image28.png
Slide 14 of 27 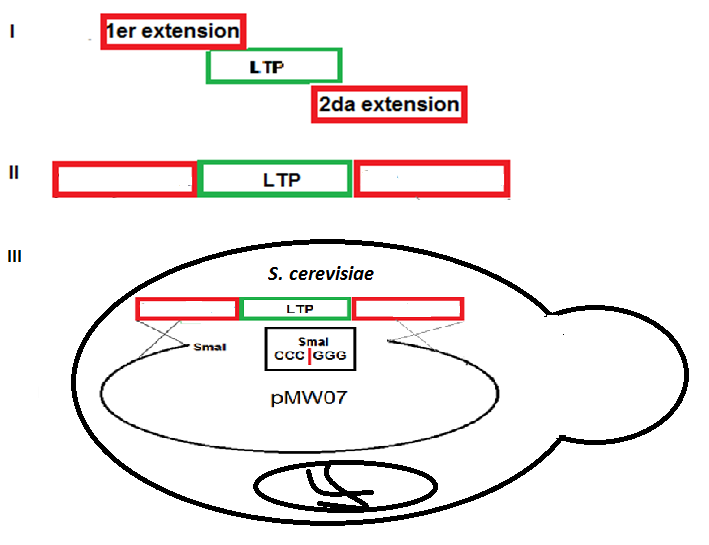
image14.png
Slide 15 of 27 
image21.png
Slide 16 of 27 
image8.png
Slide 17 of 27 
image15.png
Slide 18 of 27 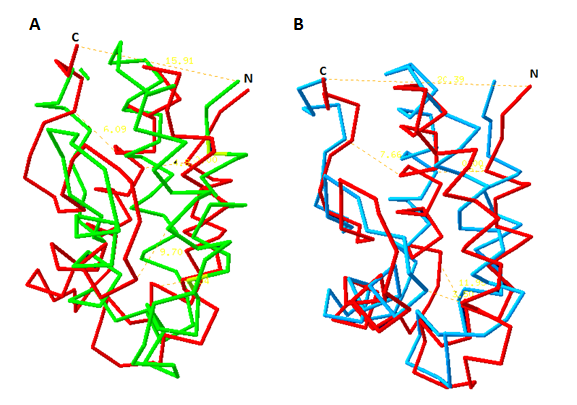
image12.png
Slide 19 of 27 
image3.png
Slide 20 of 27 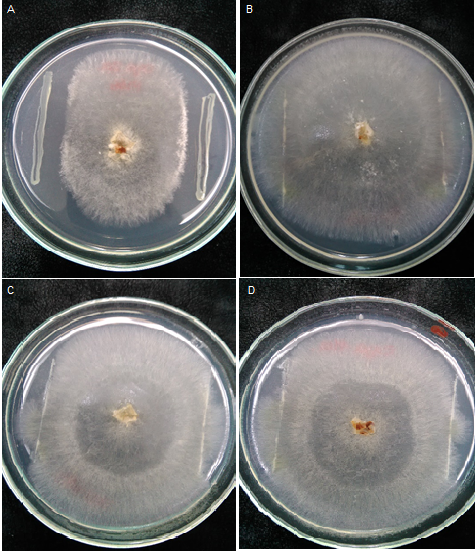
image22.png
Slide 21 of 27 
image16.png
Slide 22 of 27 
image26.png
Slide 23 of 27 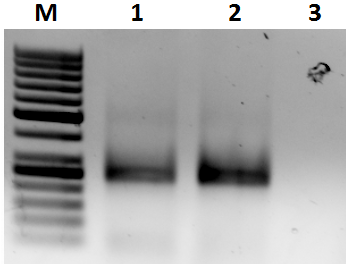
image18.png
Slide 24 of 27 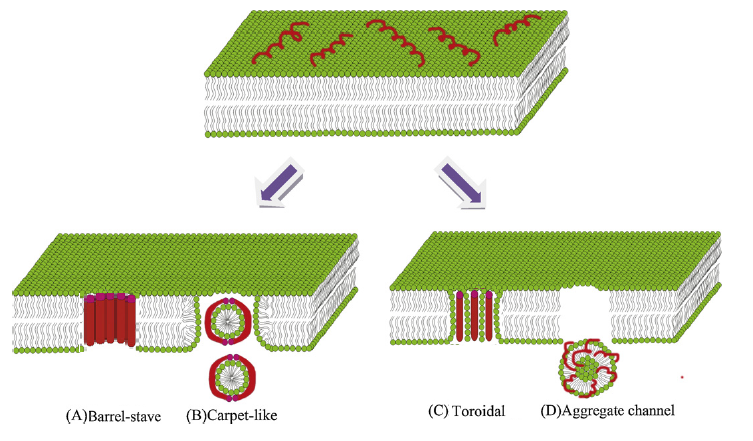
image2.png
Slide 25 of 27 
image27.png
Slide 26 of 27 
image10.png
Slide 27 of 27 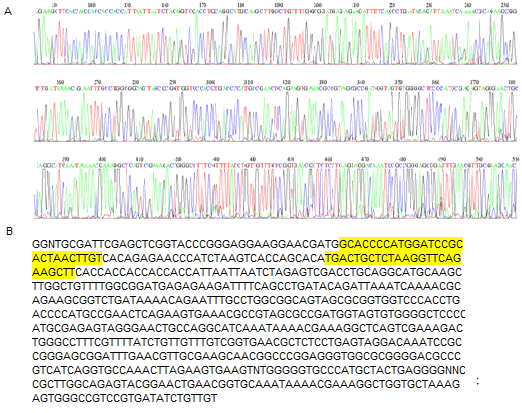
image19.png
Resumen
La manzanilla romana (Chamaemelum nobile), es una de las plantas medicinales más usadas por la presencia de flavonoides, sesquiterpenos y azulenos en sus tejidos, que le confieren propiedades antiinflamatorias, anticancerígenas y antimicrobianas. La capacidad inhibitoria de fitopatógenos de la manzanilla romana aumenta la posibilidad de identificar genes asociados con resistencia a enfermedades en plantas. Los péptidos antimicrobianos (AMPs) actúan en la inmunidad innata de plantas aumentando su presencia en tejidos susceptibles a la invasión de fitopatógenos. El objetivo de este trabajo fue identificar un posible gen relacionado a un péptido antimicrobiano a partir de Chamaemelum nobile. Se realizaron reacciones de PCR sobre ADN genómico para la identificación de génes AMPs, y se usó herramientas de Bioinformática para su caracterización bioquímica, como predicción de la secuencia de aminoácidos, punto isoeléctrico, índice alifático, hidropaticidad y modelamiento teórico de la estructura tridimensional. Luego se procedió a estandarizar el método de clonación por recombinación homóloga en levadura para AMPs, y se llevaron a cabo pruebas de sensibilidad sobre el fitopatógeno Rhizoctonia solani. Finalmente, se caracterizaron cepas para transformación genética de plantas. Se realizó la identificación de un gen AMP denominado Proteína de Transferencia de Lípidos (LTP), que posee 94 aminoácidos, a partir de material genético de C. nobile. La secuencia de aminoácidos putativa identificada en C. nobile fué caracterizada in silicio con herramientas bioinformáticas en dónde se evidenció que esta secuencia de aminoácidos es similar a la familia de Proteínas de Transferencia de Lípidos tipo 1 destacadas por presentar actividad antimicrobiana sobre fitopatógénos. Posteriormente, se evaluó el método de clonación por recombinación homóloga en levadura, y se encontró que su eficacia depende del nivel de homología vector-genoma de levadura. Adicionalmente se determinó la actividad inhibitoria de fluido apoplástico de C. nobile a 1 µg/mL sobre R. solani. Por último, se realizó la caracterización fenotípica y molecular de cepas de Agrobacterium tumefaciens que pueden ser útiles en transformación genética de plantas de interés industrial. Este estudio aporta en la descripción de un nuevo miembro de la familia de LTPs en plantas de la familia Asteraceaes.
Abstract. Roman chamomile (Chamaemelum nobile) is one of the medicinal plants most used for the presence of flavonoids, sesquiterpenes and azulenes in their tissues, which give it antiinflammatory, anticancer and antimicrobial properties. The phytopathogenic inhibitory capacity of Roman chamomile increases the possibility of identifying genes associated with disease resistance in plants. Antimicrobial peptides (AMPs) act on the innate immunity of plants by increasing their presence in tissues susceptible to phytopathogen invasion. The objective of this work was to identify a possible gene related to an antimicrobial peptide from Chamaemelum nobile. PCR reactions were performed on genomic DNA for identification of AMP genes, and Bioinformatics tools were used for biochemical characterization, such as amino acid sequence prediction, isoelectric point, aliphatic index, hydropatibility and theoretical modeling of the three-dimensional structure. Then the homologous recombination cloning method in yeast for AMPs was standardized, and sensitivity tests were performed on the phytopathogen Rhizoctonia solani. Finally, strains for genetic transformation of plants were characterized. Identification of an AMP gene called Lipid Transfer Protein (LTP), which has 94 amino acids, was made from genetic material of C. nobile. The putative amino acid sequence identified in C. nobile was characterized in silico with bioinformatic tools where it was shown that this amino acid sequence is similar to the family of Type 1 Lipid Transfer Proteins highlighted by their antimicrobial activity on phytopathogens. Subsequently, the cloning method was evaluated by homologous recombination in yeast, and it was found that its efficacy depends on the level of yeast vector-genome homology. In addition, the apoplastic fluid inhibitory activity of C. nobile at 1 μg / mL was determined on R. solani. Finally, the phenotypic and molecular characterization of Agrobacterium tumefaciens strains that may be useful in the genetic transformation of plants of industrial interest was carried out. This study contributes in the description of a new member of the family of LTPs in plants of the family Asteraceaes.
Abstract. Roman chamomile (Chamaemelum nobile) is one of the medicinal plants most used for the presence of flavonoids, sesquiterpenes and azulenes in their tissues, which give it antiinflammatory, anticancer and antimicrobial properties. The phytopathogenic inhibitory capacity of Roman chamomile increases the possibility of identifying genes associated with disease resistance in plants. Antimicrobial peptides (AMPs) act on the innate immunity of plants by increasing their presence in tissues susceptible to phytopathogen invasion. The objective of this work was to identify a possible gene related to an antimicrobial peptide from Chamaemelum nobile. PCR reactions were performed on genomic DNA for identification of AMP genes, and Bioinformatics tools were used for biochemical characterization, such as amino acid sequence prediction, isoelectric point, aliphatic index, hydropatibility and theoretical modeling of the three-dimensional structure. Then the homologous recombination cloning method in yeast for AMPs was standardized, and sensitivity tests were performed on the phytopathogen Rhizoctonia solani. Finally, strains for genetic transformation of plants were characterized. Identification of an AMP gene called Lipid Transfer Protein (LTP), which has 94 amino acids, was made from genetic material of C. nobile. The putative amino acid sequence identified in C. nobile was characterized in silico with bioinformatic tools where it was shown that this amino acid sequence is similar to the family of Type 1 Lipid Transfer Proteins highlighted by their antimicrobial activity on phytopathogens. Subsequently, the cloning method was evaluated by homologous recombination in yeast, and it was found that its efficacy depends on the level of yeast vector-genome homology. In addition, the apoplastic fluid inhibitory activity of C. nobile at 1 μg / mL was determined on R. solani. Finally, the phenotypic and molecular characterization of Agrobacterium tumefaciens strains that may be useful in the genetic transformation of plants of industrial interest was carried out. This study contributes in the description of a new member of the family of LTPs in plants of the family Asteraceaes.


